 Let’s look at some basic commands in R.
Let’s look at some basic commands in R.
Set up the following vector by cutting and pasting from this document:
a <- c(3,-7,-3,-9,3,-1,2,-12, -14)
b <- c(3,7,-5, 1, 5,-6,-9,16, -8)
d <- c(1,2,3,4,5,6,7,8,9)
Now figure out what each of the following commands do. You should not need me to explain each command, but I will explain a few. (more…)
 In Part 7, let’s look at further plotting in R. Try entering the following three commands together (the semi-colon allows you to place several commands on the same line).
In Part 7, let’s look at further plotting in R. Try entering the following three commands together (the semi-colon allows you to place several commands on the same line).
Let’s take an example with two variables and enhance it.
X <- c(3, 4, 6, 6, 7, 8, 9, 12)
B1 <- c(4, 5, 6, 7, 17, 18, 19, 22)
B2 <- c(3, 5, 8, 10, 19, 21, 22, 26)
(more…)
Item Response Theory (IRT) refers to a family of statistical models for evaluating the design and scoring of psychometric tests, assessments and surveys. It is used on assessments in psychology,  psychometrics, education, health studies, marketing, economics and social sciences — assessments that involve categorical items (e.g., Likert items).
psychometrics, education, health studies, marketing, economics and social sciences — assessments that involve categorical items (e.g., Likert items).
In this webinar, you will learn about:
- The key ideas and techniques of IRT, with examples from educational assessment
- The Rasch Model and the Graded Response Model — two of the most commonly-used IRT models
- A range of analytic techniques that should be used in conjunction with IRT
This webinar will introduce you to the basic ideas and applications of IRT, and show how you can acquire the skills necessary to conduct IRT analysis at the professional level.
Note: This training is an exclusive benefit to members of the Statistically Speaking Membership Program and part of the Stat’s Amore Trainings Series. Each Stat’s Amore Training is approximately 90 minutes long.
About the Instructor
 David Lillis is an applied statistician in Wellington, New Zealand.
David Lillis is an applied statistician in Wellington, New Zealand.
His company, Sigma Statistics and Research Limited, provides online instruction, face-to-face workshops on R, and coding services in R.
David holds a doctorate in applied statistics and is a frequent contributor to The Analysis Factor, including our blog series R is Not So Hard.
Not a Member Yet?
It’s never too early to set yourself up for successful analysis with support and training from expert statisticians.
Just head over and sign up for Statistically Speaking.
You'll get access to this training webinar, 130+ other stats trainings, a pathway to work through the trainings that you need — plus the expert guidance you need to build statistical skill with live Q&A sessions and an ask-a-mentor forum.

In Part 3 we used the lm() command to perform least squares regressions. In Part 4 we will look at more advanced aspects of regression models and see what R has to offer.
One way of checking for non-linearity in your data is to fit a polynomial model and check whether the polynomial model fits the data better than a linear model. However, you may also wish to fit a quadratic or higher model because you have reason to believe that the relationship between the variables is inherently polynomial in nature. Let’s see how to fit a quadratic model in R.
We will use a data set of counts of a variable that is decreasing over time. Cut and paste the following data into your R workspace.
A <- structure(list(Time = c(0, 1, 2, 4, 6, 8, 9, 10, 11, 12, 13,
14, 15, 16, 18, 19, 20, 21, 22, 24, 25, 26, 27, 28, 29, 30),
Counts = c(126.6, 101.8, 71.6, 101.6, 68.1, 62.9, 45.5, 41.9,
46.3, 34.1, 38.2, 41.7, 24.7, 41.5, 36.6, 19.6,
22.8, 29.6, 23.5, 15.3, 13.4, 26.8, 9.8, 18.8, 25.9, 19.3)), .Names = c("Time", "Counts"),
row.names = c(1L, 2L, 3L, 5L, 7L, 9L, 10L, 11L, 12L, 13L, 14L, 15L, 16L, 17L, 19L, 20L, 21L, 22L, 23L, 25L, 26L, 27L, 28L, 29L, 30L, 31L),
class = "data.frame")
Let’s attach the entire dataset so that we can refer to all variables directly by name.
attach(A)
names(A)
First, let’s set up a linear model, though really we should plot first and only then perform the regression.
linear.model <-lm(Counts ~ Time)
We now obtain detailed information on our regression through the summary() command.
summary(linear.model)
Call:
lm(formula = Counts ~ Time)
Residuals:
Min 1Q Median 3Q Max
-20.084 -9.875 -1.882 8.494 39.445
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 87.1550 6.0186 14.481 2.33e-13 ***
Time -2.8247 0.3318 -8.513 1.03e-08 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 15.16 on 24 degrees of freedom
Multiple R-squared: 0.7512, Adjusted R-squared: 0.7408
F-statistic: 72.47 on 1 and 24 DF, p-value: 1.033e-08
The model explains over 74% of the variance and has highly significant coefficients for the intercept and the independent variable and also a highly significant overall model p-value. However, let’s plot the counts over time and superpose our linear model.
plot(Time, Counts, pch=16, ylab = "Counts ", cex.lab = 1.3, col = "red" )
abline(lm(Counts ~ Time), col = "blue")
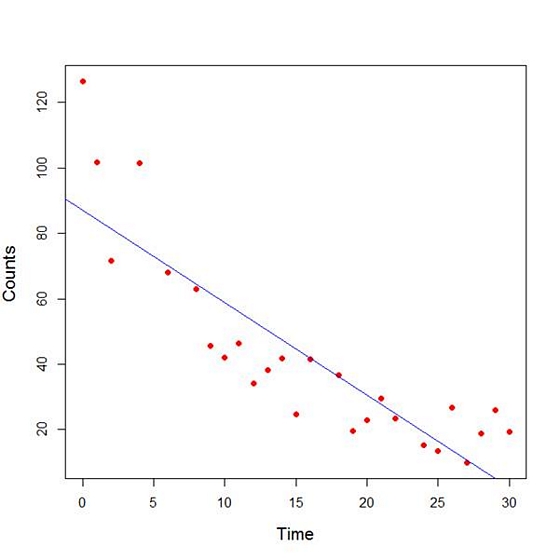
Here the syntax cex.lab = 1.3 produced axis labels of a nice size.
The model looks good, but we can see that the plot has curvature that is not explained well by a linear model. Now we fit a model that is quadratic in time. We create a variable called Time2 which is the square of the variable Time.
Time2 <- Time^2
quadratic.model <-lm(Counts ~ Time + Time2)
Note the syntax involved in fitting a linear model with two or more predictors. We include each predictor and put a plus sign between them.
summary(quadratic.model)
Call:
lm(formula = Counts ~ Time + Time2)
Residuals:
Min 1Q Median 3Q Max
-24.2649 -4.9206 -0.9519 5.5860 18.7728
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 110.10749 5.48026 20.092 4.38e-16 ***
Time -7.42253 0.80583 -9.211 3.52e-09 ***
Time2 0.15061 0.02545 5.917 4.95e-06 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 9.754 on 23 degrees of freedom
Multiple R-squared: 0.9014, Adjusted R-squared: 0.8928
F-statistic: 105.1 on 2 and 23 DF, p-value: 2.701e-12
Our quadratic model is essentially a linear model in two variables, one of which is the square of the other. We see that however good the linear model was, a quadratic model performs even better, explaining an additional 15% of the variance. Now let’s plot the quadratic model by setting up a grid of time values running from 0 to 30 seconds in increments of 0.1s.
timevalues <- seq(0, 30, 0.1)
predictedcounts <- predict(quadratic.model,list(Time=timevalues, Time2=timevalues^2))
plot(Time, Counts, pch=16, xlab = "Time (s)", ylab = "Counts", cex.lab = 1.3, col = "blue")
Now we include the quadratic model to the plot using the lines() command.
lines(timevalues, predictedcounts, col = "darkgreen", lwd = 3)
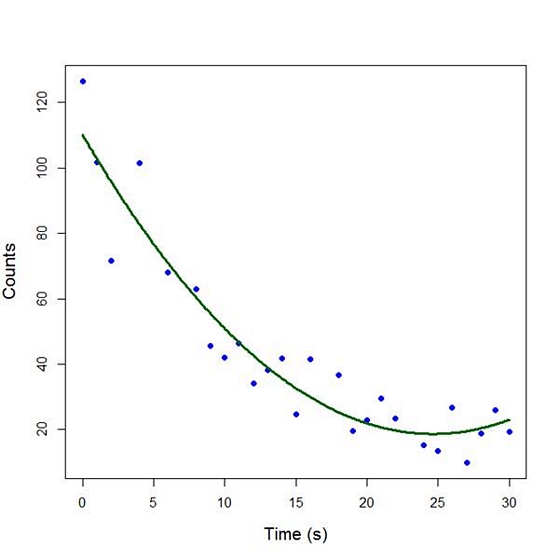
The quadratic model appears to fit the data better than the linear model. We will look again at fitting curved models in our next blog post.
About the Author: David Lillis has taught R to many researchers and statisticians. His company, Sigma Statistics and Research Limited, provides both on-line instruction and face-to-face workshops on R, and coding services in R. David holds a doctorate in applied statistics.
See our full R Tutorial Series and other blog posts regarding R programming.

We were recently fortunate to host a free The Craft of Statistical Analysis Webinar with guest presenter David Lillis. As usual, we had hundreds of attendees and didn’t get through all the questions. So David has graciously agreed to answer questions here.
If you missed the live webinar, you can download the recording here: Ten Data Analysis Tips in R.
Q: Is the M=structure(.list(.., class = “data.frame) the same as M=data.frame(..)? Is there some reason to prefer to use structure(list, … ,) as opposed to M=data.frame?
A: They are not the same. The structure( .. .) syntax is a short-hand way of storing a data set. If you have a data set called M, then the command dput(M) provides a shorthand way of storing the dataset. You can then reconstitute it later as follows: M <- structure( . . . .). Try it for yourselves on a rectangular dataset. For example, start off with (more…)
 Let’s look at some basic commands in R.
Let’s look at some basic commands in R.

 David Lillis is an applied statistician in Wellington, New Zealand.
David Lillis is an applied statistician in Wellington, New Zealand.

